Metabolomics Developing Core Facility
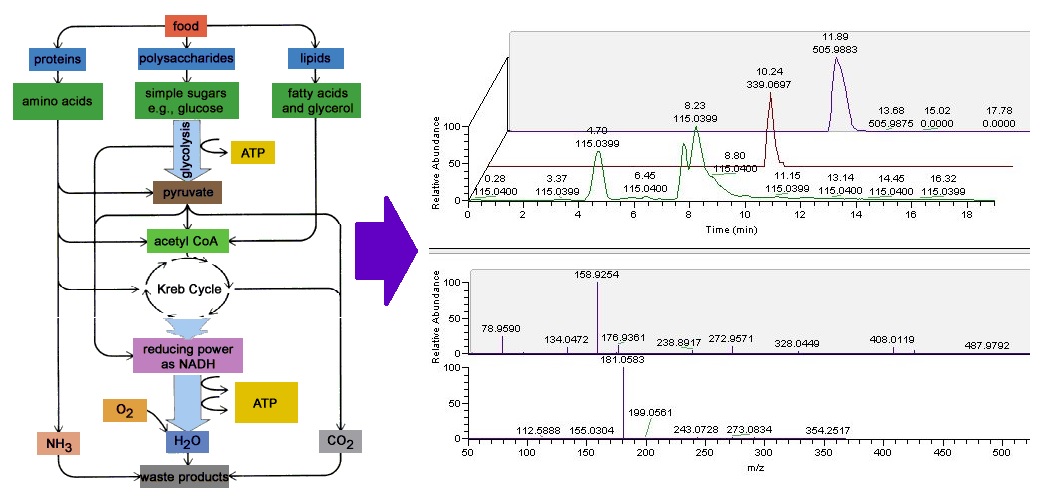
The mission of Metabolomics Core Facility (MCF) at Robert H. Lurie Comprehensive Cancer Center of Northwestern University is to provide: 1. LCMS based metabolomics service including identification, semi-quantification and quantification of metabolites from biological samples including but not limited to cells, bio-fluids and tissues; 2. integrative genomics service related to metabolism. MCF supports investigators engaged in basic, preclinical and clinical cancer research, including scientists examining basic mechanisms in of disease, as well as those seeking to identify novel targets for therapy or biomarkers that can be used for early detection, diagnosis, prognosis or response to therapy.
Contact Us
Metabolomics
- Olson 8-310
710 N. Fairbanks Court
Chicago, IL 60611 - 312-908-8312
- metabo@northwestern.edu
Integrative Genomics
- Simpson Querrey 5-312
303 E Superior St.
Chicago, IL 60611 - 312-503-0534
- Metabo.Genomics@northwestern.edu
Services and FAQs
Core Introduction & Metabolomics 101
Key Services
- Profiling - Comprehensive Hydrophilic Metabolites Panel (CHMP) - LCMS Run
Use LCMS to acquire signals from a broad spectrum of water soluble metabolites (250 plus targets including but not limited to amino acids, nucleotides and nucleobase/nucleosides, cofactors, Krebs cycle, TCA cycle, urea cycle, glycolysis, gluconeogenesis, pentose phosphate pathway, etc. See Target list for details). Semi-quantitative (comparison between samples) 30-40 minute run with raw data available. Fee/injection. - Profiling - Targeted Metabolites (PTM) - LCMS Run
Use LCMS to acquire signals for specific targets-of-your-interest (hydrophilic metabolites or lipids). Semi-quantitative (comparison between samples) 25-35 minute run with raw data available. Fee/injection. - Profiling - Fluxomics (FLUX) - LCMS Run
Use LCMS to acquire signals from specific stable isotopes labeled metabolites generated through incubation of tracers, such as 13C, 15N, 2H to study metabolic flux in specific pathways. Semi-quantitative (comparison between samples) 25-35 minute LCMS run with raw data available. Fee/injection. - Quantification – Targeted Metabolites (QTM) - LCMS Run
Use LCMS to quantify targets-of-your-interest (hydrophilic metabolites or lipids) from sample extractions. Internal standards and standard compounds for targets are required. Fee/injection. - Untargeted Metabolomics – Hydrophilic Metabolites (UMHM) – LCMS Run
Use LCMS to acquire signals from water soluble metabolites extractions. Raw data from untargeted metabolomics method (full MS1 scan with data dependent MS2) will be provided. Researchers will choose their own choice of partners (including core’s collaborator from outside institute) to perform untargeted metabolomics analysis. Fee/injection. - Sample Preparation
Process samples for LCMS injection. Fee/hour. - Data (LCMS) analysis
For profiling services, targets will be annotated, and peak areas (artificial unit) will be reported for semi-quantification (comparison between samples). For quantification services, amount or concentration will be reported. Fee/hour. For untargeted metabolomics service, raw data will be provided, and researchers will choose their own choice of partners (including core’s collaborator from outside institute) to perform untargeted metabolomics analysis. - Method Development
Develop new assay per customer request. - SeaHorse Analyzers
Shared instruments will be provided for researchers to perform self-guided SeaHorse experiments. Fee/hour. - Consulting
Sample Submission Process
- Please contact the correct core branch to discuss the project and submission.
- Once ready to submit samples, visit https://nucore.northwestern.edu/facilities/Metabo and follow the instruction to generate an order.
- Print out the filled Sample Submission Form generated by NUCore ordering and bring samples to core.
FAQs
- Starting Materials - It is important to keep the amount of starting material (cell number, equivalent protein amount, bio-liquid volume and tissue weight, etc.) as same as possible across the batch. Starting material amount is required during submission to calculate injection amount.
- Standard Compounds - For quantification services, internal standards and standard compounds for targets are required. For profiling services, standard compounds are recommend if certain metabolites are the targets-of-interest.
- Turnaround Time - Under normal circumstances, 10 business days are typical for medium-low complexity projects. Projects that are more complex take longer.
- Sample/Data Storage - Leftover samples (if any) will be kept in -20°C freezer for 3 months, and the raw data will be kept for up to 12 months.
- Approved price list (please contact core)
Equipment
- UHPLC-MS (Hybrid Quadrupole-Orbitrap): Thermo Ultimate-3000 + Q-exactive
- UHPLC-MS (Triple Quadrupole): Thermo Vanquish + TSQ
- SeaHorse Analyzer: XFe96 – 96-well plate
- SeaHorse Analyzer: XFp – 8-well plate
- Speed-Vac
Selected Publications
- mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis. Mol Cell. 2021 Mar 22. doi.org/10.1016/j.molcel.2021.03.009.
- Polyamines drive myeloid cell survival by buffering intracellular pH to promote immunosuppression in glioblastoma. Sci Adv. 2021 Feb 17;7(8):eabc8929. doi: 10.1126/sciadv.abc8929.
- Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature. 2020 Sep;585(7824):288-292. doi: 10.1038/s41586-020-2475-6.
- ERK2 Phosphorylates PFAS to Mediate Posttranslational Control of De Novo Purine Synthesis. Mol Cell. 2020 Jun 18;78(6):1178-1191.e6. doi: 10.1016/j.molcel.2020.05.001.
- Direct stimulation of NADP+ synthesis through Akt-mediated phosphorylation of NAD kinase. Science. 2019 Mar 8;363(6431):1088-1092. doi: 10.1126/science.aau3903.
- Metformin targets mitochondrial electron transport to reduce air-pollution-induced thrombosis. Cell Metab. 2019 Feb 5;29(2):335-347.e5. doi: 10.1016/j.cmet.2018.09.019.
Acknowledgement
Work performed in Metabolomics Core Facility should be acknowledged as following:
- Metabolomics services were performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University.



